PDB-Repair
Using PDB-Repair to fix atomic missing on the main/side chain of the input protein file
1. Upload your PDB files that need to be fixed, the page will check whether the uploaded files meet the requirements
2. When the input protein PDB file has only missing atoms and no missing residues, the program can directly perform the repair: when a single main-chain atom is missing, the program can directly compute the repair; When the main chain atoms are missing two or more, we consider this amino acid to be missing, the program use CCD algorithm to complete it.
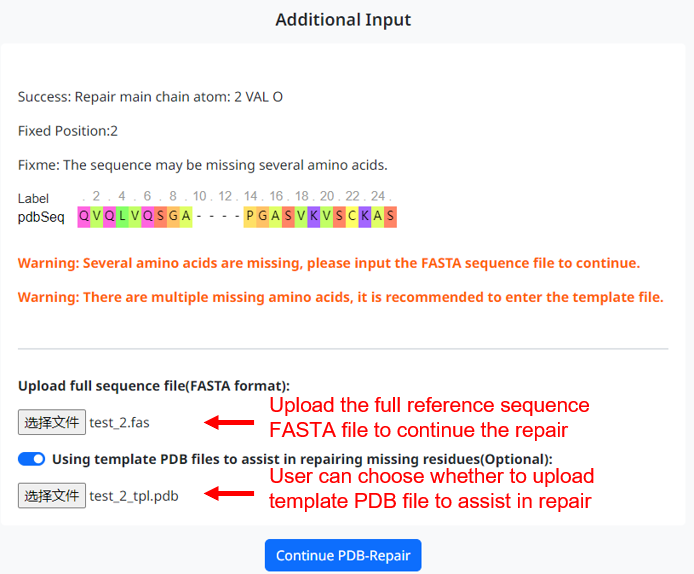
3. If the PDB file you upload is missing several residues, the user needs to provide a complete reference sequence FASTA file for the program to continue the repair. If multiple residues are missing, it is recommended that the user select the option to provide a template PDB file. This allows the program to match the missing regions with the corresponding template structure, making the repaired structure more reasonable.
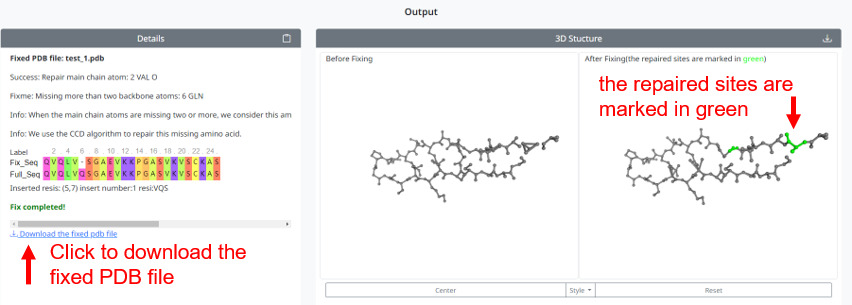
4. When the repair is complete, the page shows the details of the repair and the 3D structure of the PDB file before and after the repair. Also provide the file download after repair
Canutescu A A, Dunbrack Jr R L. Cyclic coordinate descent: A robotics algorithm for protein loop closure[J]. Protein science, 2003, 12(5): 963-972.