AA-Mutate
Using AA-mutate to generate the mutated conformation of the selected residue on the input protein.The 3D structure before and after the mutation will be displayed on the page. AA-Mutate is based on side-chain modeling method CIS-RR.
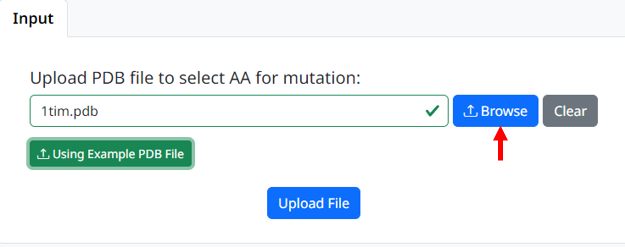
1. Upload your PDB files that need to be mutated, the page will check whether the uploaded file meets the requirements
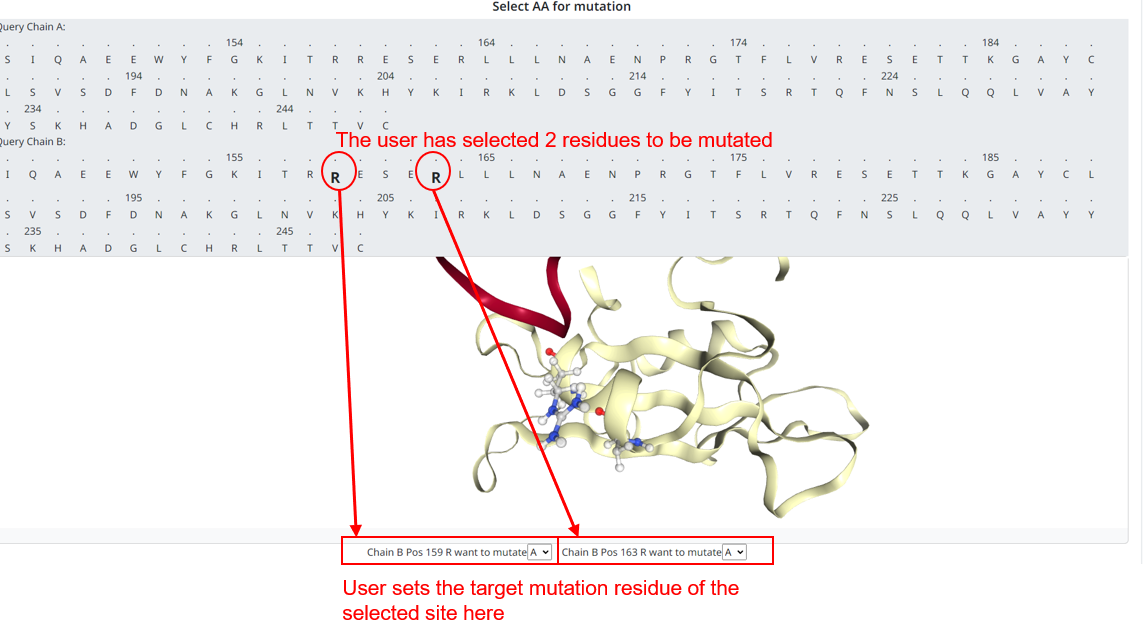
2. The program will analyze the uploaded PDB file and provide a secondary input interface for mutation selection. In this page, all the mutageable sites in the chain will be displayed, and the user can click on the site that wants to mutate to select (after selecting the site will be enlarged display) . After selection, the position and structure of the residue is indicated in the 3D structure, and the mutation target input box appears below the 3D box, where the user enters which amino acid he wants the site to be mutated to. The user can click the selected residue again to deselect it
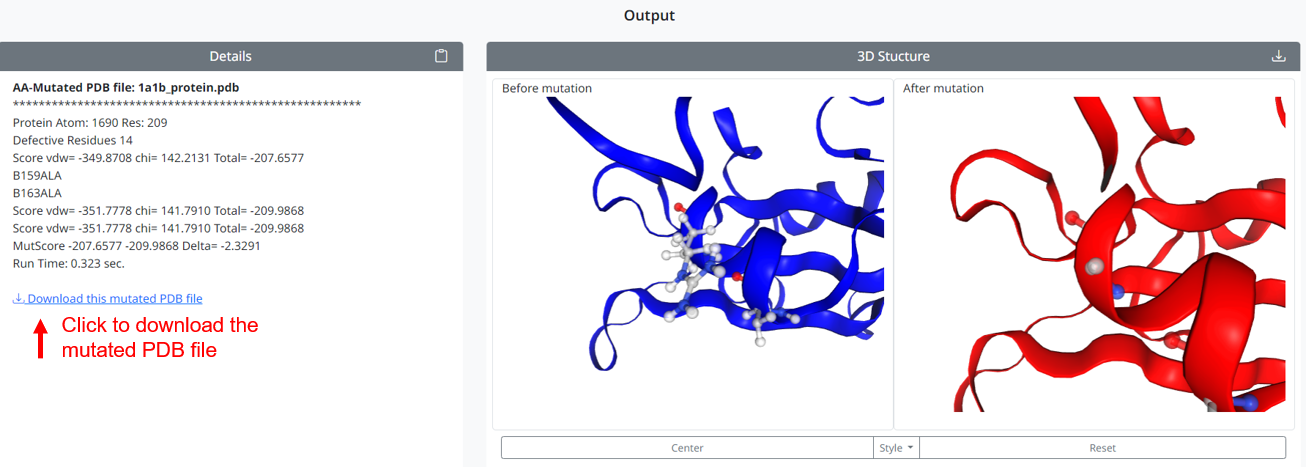
3. After the user has selected and typed, click the 'Run AA-Mutate' button. The program outputs the results: the details of the mutation are shown in the left box, and the positions and structures of the selected residues before and after mutation are shown in the right 3D box. Users can click on the link to download the mutated PDB file
Cao Y, Song L, Miao Z, et al. Improved side-chain modeling by coupling clash-detection guided iterative search with rotamer relaxation[J]. Bioinformatics, 2011, 27(6): 785-790.