StuctureAlign
Using StructureAlign to Superpose two proteins to evaluate their structural similarity (support domain alignment)
1. Upload your query PDB and reference PDB files that need to be aligned, the page will check whether the uploaded files meet the requirements
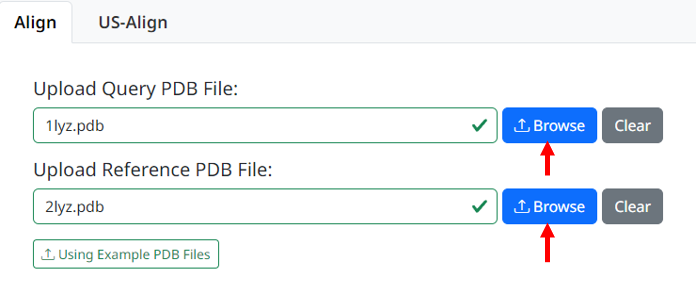
2. After clicking the 'Upload Files' button, you have the option of global alignment (the default) or custom range alignment. If you want to perform global alignment, there is no need to input additional parameters. Simply click the Run StructureAlign button to continue
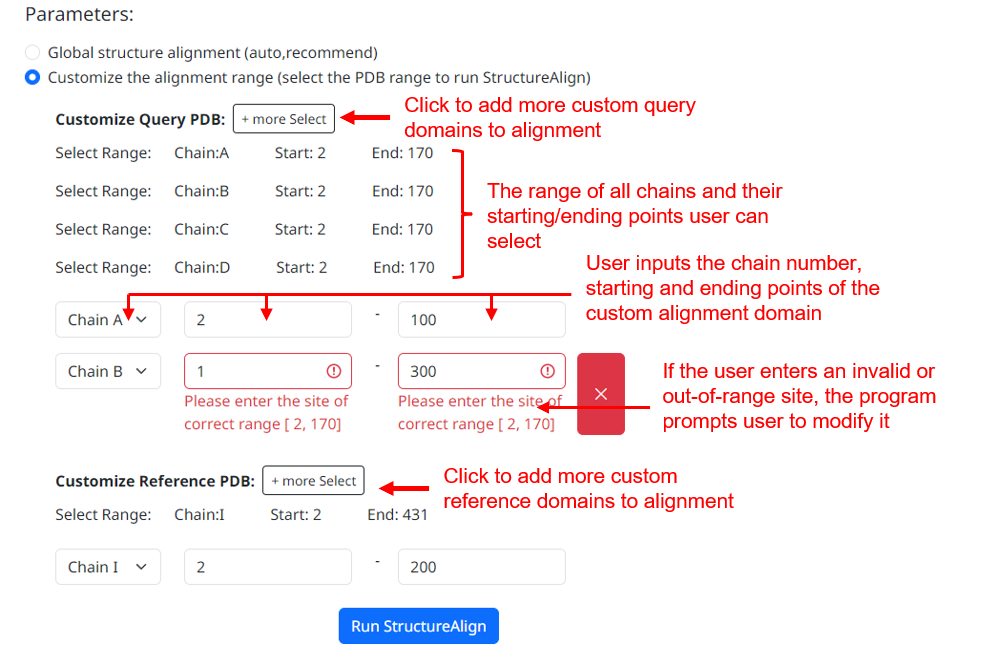
3. If you choose to do a custom range alignment, the page will analyze the PDB file you uploaded and give the custom range parameter input interface. You can select any number of truncated regions for alignment. The page displays a customizable range of chains and starting and ending points that the user's input needs to meet.
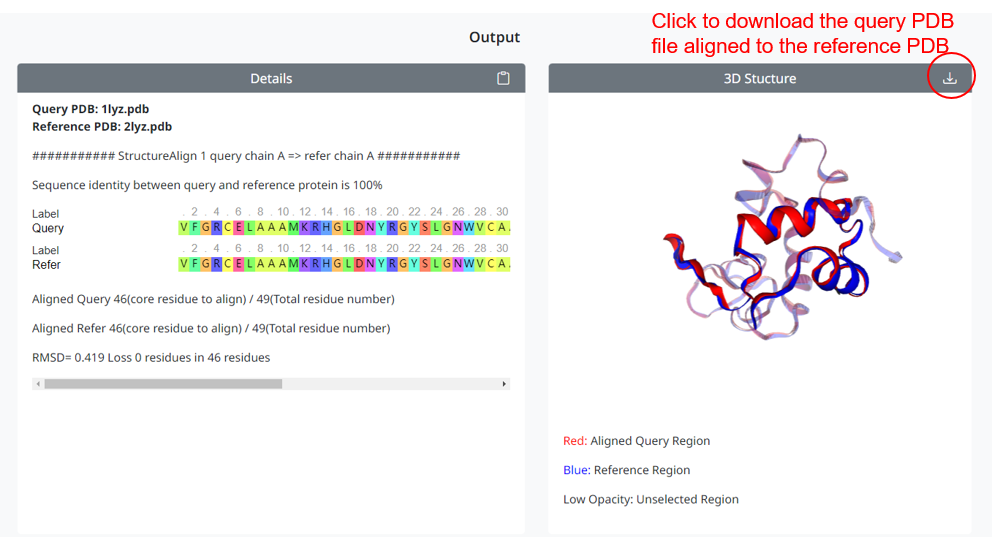
4. After the user inputs correctly and clicks the Run StructureAlign button to submit, the alignment result will be displayed on the webpage. The left box shows the alignment details, including sequence alignment and RMSD value. The 3D conformation overlaid with the query and reference PDB is shown in the box on the right. If you select custom range alignment, the non custom range will be displayed semi transparently. You can click on the download icon to download the aligned query conformation.
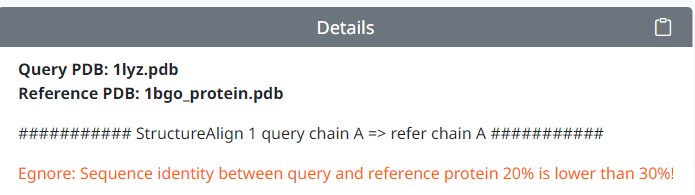
5. If your input alignment range sequence similarity is less than 30% , the program will be difficult to give an accurate alignment conformations, will give the following error message. At this point we recommend that you try US-align instead of align, see 'US-align Tutorial' for more help
Using US-align for global protein alignment (no sequence similarity requirement)
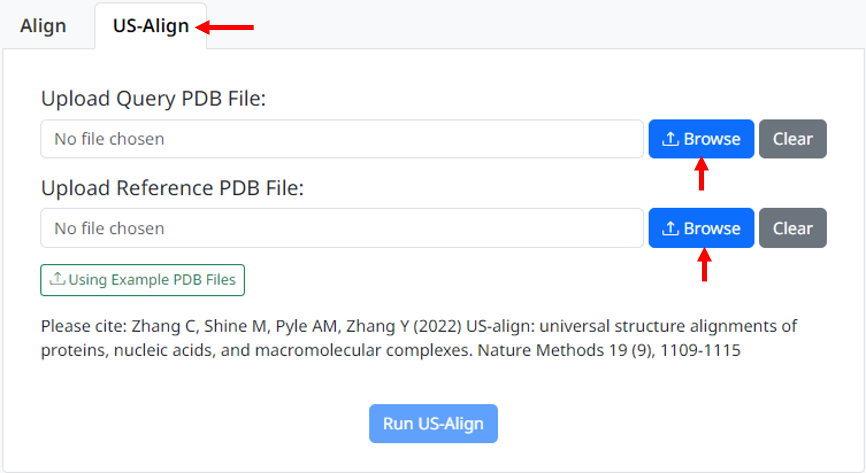
1. Click on the US-align tab box on the right of align to enter the US-align page. Upload your query PDB and reference PDB files that need to be aligned, the page will check whether the uploaded files meet the requirements
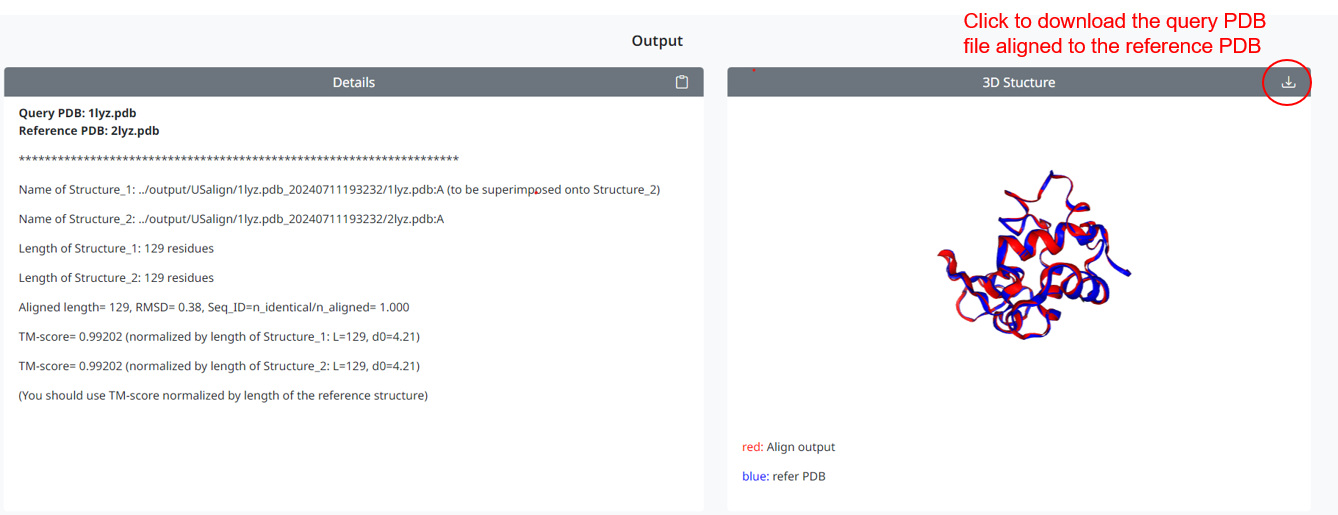
2. When the user clicks the Run US-align button to submit, the page displays the alignment results: in the left box are the alignment details, including the RMSD value and TM-Score; in the right box are the 3D conformations of the query superimposed on the reference sequence. You can click the download icon to download the query conformations superimposed on the reference PDB conformations.
Zhang C, Shine M, Pyle A M, et al. US-align: universal structure alignments of proteins, nucleic acids, and macromolecular complexes[J]. Nature methods, 2022, 19(9): 1109-1115.